Tools for Analysis and Visualization of Large Complex Data in R
Rencontres R
June 23, 2016
Follow along live: http://bit.ly/rr2015hafenv
View at your own pace: http://bit.ly/rr2015hafen
Packages to Install
install.packages(c("datadr", "trelliscope", "rbokeh"))(If you want to try it out - not required)
library(trelliscope)
library(rbokeh)
library(housingData)
panel <- function(x)
figure() %>% ly_points(time, medListPriceSqft, data = x)
housing %>%
qtrellis(by = c("county", "state"), panel, layout = c(2, 4))
# note: this will open up a web browser with a shiny app running
# use ctrl-c or esc to get back to your R promptTo test the installation:
Background
Deep Analysis of Large Complex Data
Deep Analysis of
Large Complex Data
-
Data most often does not come with a model
-
If we already (think we) know the algorithm / model to apply and simply apply it to the data and do nothing else to assess validity, we are not doing analysis, we are processing
-
Deep analysis means detailed, comprehensive analysis that does not lose important information in the data
-
It means trial and error, an iterative process of hypothesizing, fitting, validating, learning
-
It means a lot of visualization
-
It means access to 1000s of statistical, machine learning, and visualization methods, regardless of data size
-
It means access to a high-level language to make iteration efficient
R is Great for Deep Analysis
-
Rapid prototyping
-
Access to 1000s of methods including excellent statistical visualization tools
-
Data structures and idioms tailored for data analysis
But R struggles with big data...
Deep Analysis of
Large Complex Data
Any or all of the following:
- Large number of records
- Many variables
- Complex data structures not readily put into tabular form of cases by variables
- Intricate patterns and dependencies that require complex models and methods of analysis
- Not i.i.d.!
Often complex data is more of a challenge than large data, but most large data sets are also complex
Goals
- Stay in R regardless of size of data
- Be able to use all methods available in R regardless of size of data
- Minimize both computation time (through scaling) and analyst time (through a simple interface in R) - with emphasis on analyst time
- Flexible data structures
- Scalable while keeping the same simple interface
Approach
Methodology:
Divide and Recombine
Software Implementation:
Tessera
Divide and Recombine
Divide and Recombine
- Specify meaningful, persistent partioning of the data
- Analytic or visual methods are applied independently to each subset of the divided data in embarrassingly parallel fashion
- Results are recombined to yield a statistically valid D&R result for the analytic method
Simple Idea
Divide and Recombine
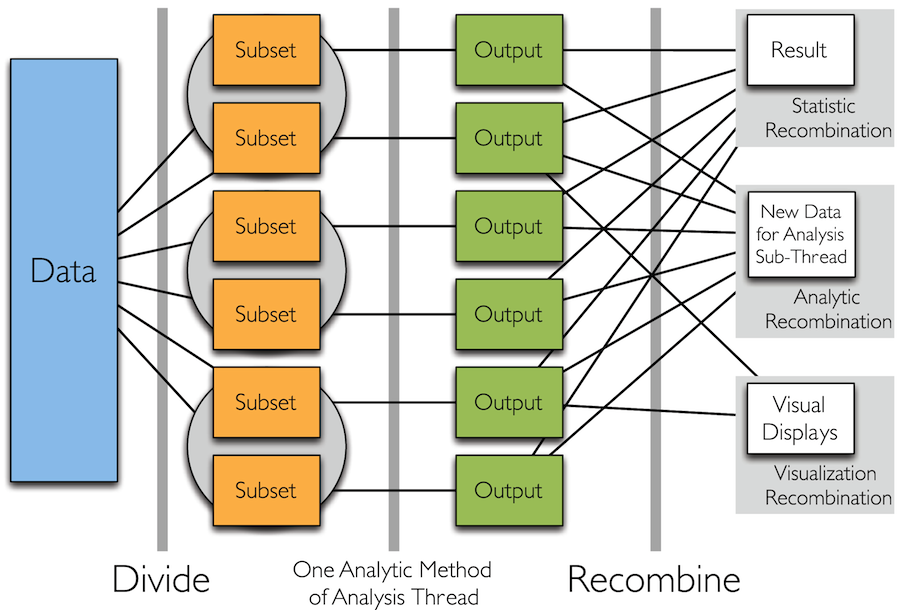
How to Divide the Data?
- Typically "big data" is big because it is made up of collections of smaller data from many subjects, sensors, locations, time periods, etc.
- It is therefore natural to break the data up based on these variables
- We call this conditioning variable division
- It is in practice by far the most common thing we do (and it's nothing new)
- Another option is random replicate division

Analytic Recombination
- Analytic recombination begins with applying an analytic method independently to each subset
- We can use any of the small-data methods we have available (think of the 1000s of methods in R)
- For conditioning-variable division:
- Typically the recombination depends on the subject matter
- Example: fit a linear model to each subset and combine the estimated coefficients for each subset to build a statistical model or for visual study

Analytic Recombination
- For random replicate division:
- Division methods are based on statistical matters, not the subject matter as in conditioning-variable division
- Results are often approximations
- Approaches that fit this paradigm
- Coefficient averaging
- Subset likelihood modeling
- Bag of little bootstraps
- Consensus MCMC

Visual Recombination
-
Most visualization tools & approaches for big data either
- Summarize lot of data to create a single plot
- Are very specialized for a particular domain
- Summaries are critical
- But we must be able to flexibly visualize complex data in detail even when they are large!
Trellis Display
- Data are split into meaningful subsets, usually conditioning on variables of the dataset
- A visualization method is applied to each subset
- The image for each subset is called a "panel"
- Panels are arranged in an array of rows, columns, and pages, resembling a garden trellis
Why Trellis is Effective
- Easy and flexible to create
- Data complexity / dimensionality can be handled by splitting the data up
- We have a great deal of freedom with what we plot inside each panel
- Easy and effective to consume
- Once a viewer understands one panel, they have immediate access to the data in all other panels
- Scanning across panels elicits comparisons to reveal repetition and change, pattern and surprise

Scaling Trellis
- Big data lends itself nicely to the idea of Trellis Display
- Recall that typically "big data" is big because it is made up of collections of smaller data from many subjects, sensors, locations, time periods, etc.
- It is natural to break the data up based on these dimensions and make a plot for each subset
- But this could mean Trellis displays with potentially thousands or millions of panels
- We can create millions of plots, but we will never be able to (or want to) view all of them!
Scaling Trellis
-
To scale, we can apply the same steps as in Trellis display, with one extra step:
- Data are split into meaningful subsets, usually conditioning on variables of the dataset
- A visualization method is applied to each subset
- A set of cognostics that measure attributes of interest for each subset is computed
- Panels are arranged in an array of rows, columns, and pages, resembling a garden trellis, with the arrangement being specified through interactions with the cognostics
Trelliscope is Scalable
- 6 months of US high frequency equity trading data
- Hundreds of gigabytes of data
- Split by stock symbol and day
- Nearly 1 million subsets
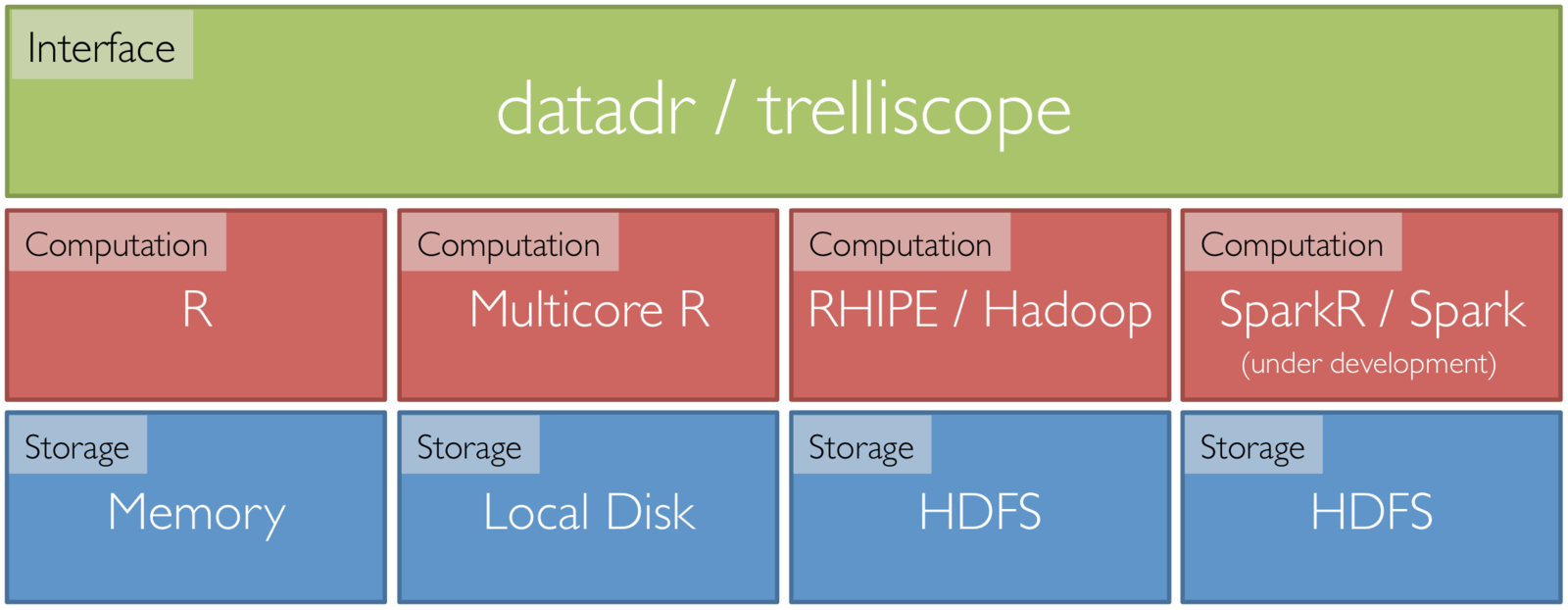
Tessera
An Implementation of D&R in R
Tessera
Front end R packages that can tie to scalable back ends:
- datadr: an interface to data operations, division, and analytical recombination methods
- trelliscope: visual recombination through interactive multipanel exploration with cognostics
Data Structures in datadr
-
Must be able to break data down into pieces for independent storage / computation
-
Recall the potential for: “Complex data structures not readily put into tabular form of cases by variables”
-
Key-value pairs: a flexible storage paradigm for partitioned, distributed data
-
each subset of a division is an R list with two elements: key, value
-
keys and values can be any R object
-
# bySpecies <- divide(iris, by = "Species")
[[1]]
$key
[1] "setosa"
$value
Sepal.Length Sepal.Width Petal.Length Petal.Width
5.1 3.5 1.4 0.2
4.9 3.0 1.4 0.2
4.7 3.2 1.3 0.2
4.6 3.1 1.5 0.2
5.0 3.6 1.4 0.2
...
[[2]]
$key
[1] "versicolor"
$value
Sepal.Length Sepal.Width Petal.Length Petal.Width
7.0 3.2 4.7 1.4
6.4 3.2 4.5 1.5
6.9 3.1 4.9 1.5
5.5 2.3 4.0 1.3
6.5 2.8 4.6 1.5
...
[[3]]
$key
[1] "virginica"
$value
Sepal.Length Sepal.Width Petal.Length Petal.Width
6.3 3.3 6.0 2.5
5.8 2.7 5.1 1.9
7.1 3.0 5.9 2.1
6.3 2.9 5.6 1.8
6.5 3.0 5.8 2.2
...Example: Fisher's iris data
We can represent the data divided by species as a collection of key-value pairs
- Key is species name
- Value is a data frame of measurements for that species
Measurements in centimeters of the variables sepal length and width and petal length and width for 50 flowers from each of 3 species of iris: "setosa", "versicolor", and "virginica"
Each key-value pair is an independent unit and can be stored across disks, machines, etc.
[[1]]
$key
[1] "setosa"
$value
Sepal.Length Sepal.Width Petal.Length Petal.Width
5.1 3.5 1.4 0.2
4.9 3.0 1.4 0.2
4.7 3.2 1.3 0.2
4.6 3.1 1.5 0.2
5.0 3.6 1.4 0.2
...
[[2]]
$key
[1] "versicolor"
$value
Sepal.Length Sepal.Width Petal.Length Petal.Width
7.0 3.2 4.7 1.4
6.4 3.2 4.5 1.5
6.9 3.1 4.9 1.5
5.5 2.3 4.0 1.3
6.5 2.8 4.6 1.5
...
[[3]]
$key
[1] "virginica"
$value
Sepal.Length Sepal.Width Petal.Length Petal.Width
6.3 3.3 6.0 2.5
5.8 2.7 5.1 1.9
7.1 3.0 5.9 2.1
6.3 2.9 5.6 1.8
6.5 3.0 5.8 2.2
...Distributed Data Objects &
Distributed Data Frames
A collection of key-value pairs that forms a data set is called a distributed data object (ddo) and is the fundamental data type that is passed to and returned from most of the functions in datadr.
A ddo for which every value in each key value pair is a data frame of the same schema is called a distributed data frame (ddf)
This is a ddf (and a ddo)
[[1]]
$key
[1] "setosa"
$value
Call:
loess(formula = Sepal.Length ~ Sepal.Width, data = x)
Number of Observations: 50
Equivalent Number of Parameters: 5.53
Residual Standard Error: 0.2437
[[2]]
$key
[1] "versicolor"
$value
Call:
loess(formula = Sepal.Length ~ Sepal.Width, data = x)
Number of Observations: 50
Equivalent Number of Parameters: 5
Residual Standard Error: 0.4483
[[3]]
$key
[1] "virginica"
$value
Call:
loess(formula = Sepal.Length ~ Sepal.Width, data = x)
Number of Observations: 50
Equivalent Number of Parameters: 5.59
Residual Standard Error: 0.5683 Distributed Data Objects &
Distributed Data Frames
Suppose we fit a loess model to each subset and store the result in a new collection of key-value pairs.
This is a ddo but not a ddf
datadr Data Connections
-
Distributed data types / backend connections
-
localDiskConn(), hdfsConn() connections to ddo / ddf objects persisted on a backend storage system
-
ddo(): instantiate a ddo from a backend connection
-
ddf(): instantiate a ddf from a backend connection
-
-
Conversion methods between data stored on different backends

Computation in datadr
MapReduce
-
A general-purpose paradigm for processing big data sets
-
The standard processing paradigm in Hadoop
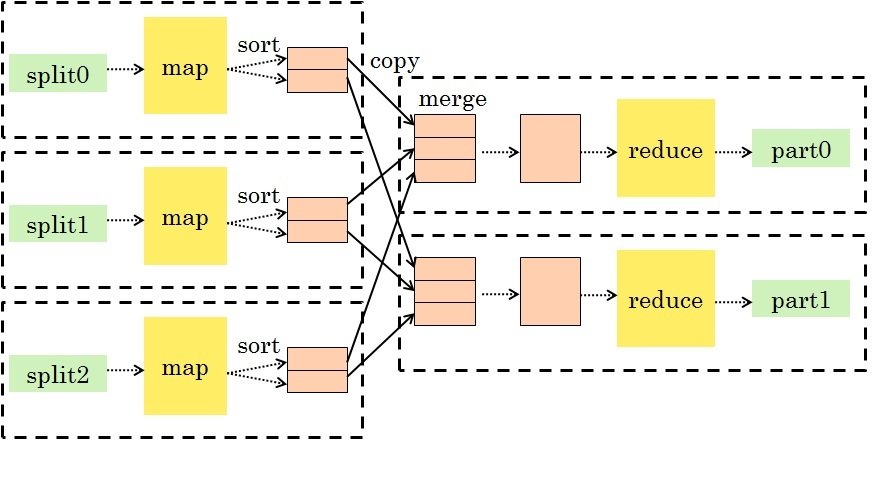
MapReduce
- Map: take input key-value pairs and apply a map function, emitting intermediate transformed key-value pairs
-
Shuffle: group map outputs by intermediate key
-
Reduce: for each unique intermediate key, gather all values and process with a reduce function

This is a very general and scalable approach to processing data.
MapReduce Example
- Map: for each species in the input key-value pair, emit the species as the key and the max as the value
- Reduce: for each grouped intermediate species key, compute the maximum of the values
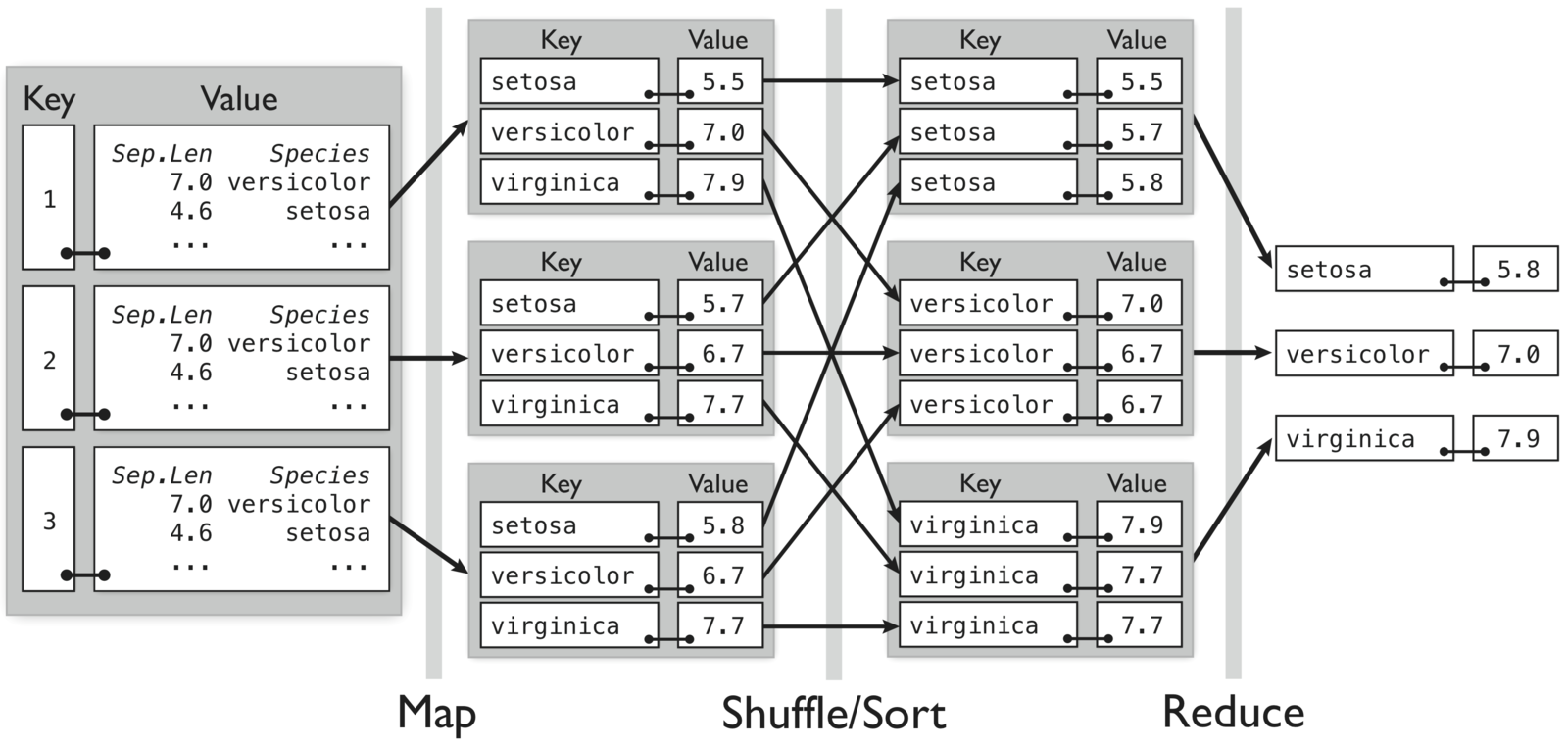
Compute maximum sepal length by species for randomly divided iris data
MapReduce Example
maxMap <- expression({
# map.values: list of input values
for(v in map.values) {
# v is a data.frame
# for each species, "collect" a
# k/v pair to be sent to reduce
by(v, v$Species, function(x) {
collect(
x$Species[1], # key
max(x$Sepal.Length) # value
)
})
}
})Map
# reduce is called for unique intermediate key
# "pre" is an initialization called once
# "reduce" is called for batches of reduce.values
# "post" runs after processing all reduce.values
maxReduce <- expression(
pre={
speciesMax <- NULL
},
reduce={
curValues <- do.call(c, reduce.values)
speciesMax <- max(c(speciesMax, curValues))
},
post={
collect(reduce.key, speciesMax)
}
)Reduce
This may not look very intuitive
But could be run on hundreds of terabytes of data
And it's much easier than what you'd write in Java
Computation in datadr
- Everything uses MapReduce under the hood
-
As often as possible, functions exist in datadr to shield the analyst from having to write MapReduce code
MapReduce is sufficient
for all D&R operations
D&R methods in datadr
-
A divide() function takes a ddf and splits it using conditioning or random replicate division
-
Analytic methods are applied to a ddo/ddf with the addTransform() function
-
Recombinations are specified with recombine(), which provides some standard combiner methods, such as combRbind(), which binds transformed results into single data frame
-
Division of ddos with arbitrary data structures must typically be done with custom MapReduce code (unless data can be temporarily transformed into a ddf)
datadr Other Data Operations
-
drLapply(): apply a function to each subset of a ddo/ddf and obtain a new ddo/ddf
-
drJoin(): join multiple ddo/ddf objects by key
-
drSample(): take a random sample of subsets of a ddo/ddf
-
drFilter(): filter out subsets of a ddo/ddf that do not meet a specified criteria
-
drSubset(): return a subset data frame of a ddf
-
drRead.table() and friends
-
mrExec(): run a traditional MapReduce job on a ddo/ddf
datadr Division Independent Methods
-
drQuantile(): estimate all-data quantiles, optionally by a grouping variable
-
drAggregate(): all-data tabulation
-
drHexbin(): all-data hexagonal binning aggregation
-
summary() method computes numerically stable moments, other summary stats (freq table, range, #NA, etc.)
-
More to come
Scalable Setup
Set up your own Hadoop cluster:
http://tessera.io/docs-install-cluster/
Set up a cluster on Amazon Web Services:
./tessera-emr.sh -n 10 -s s3://tessera-emr -m m3.xlarge -w m3.xlarge
What's Different About Tessera?
- Restrictive in data structures (only data frames / tabular / matrix algebra)
- Restrictive in methods (only SQL-like operations or a handful of scalable non-native algorithms)
- Or both!
Many other "big data" systems that support R are either:
Idea of Tessera:
- Let's use R for the flexibility it was designed for, regardless of the size of the data
- Use any R data structure and run any R code at scale
More Information
- website: http://tessera.io
- code: http://github.com/tesseradata
- Twitter: @TesseraIO
- Google user group
-
Try it out
- If you have some applications in mind, give it a try!
- You don’t need big data or a cluster to use Tessera
- Ask us for help, provide feedback, join in
For more information (docs, code, papers, user group, blog, etc.): http://tessera.io
Tangent 1: http://hafen.github.io/rbokeh/
Tangent 2: http://gallery.htmlwidgets.org/
Thank You
Small Data Example You Can Experiment With
US Home Prices by State and County from Zillow
Zillow Housing Data
- Housing sales and listing data in the United States
- Between 2008-10-01 and 2014-03-01
- Aggregated to the county level
- Zillow.com data provided by Quandl
library(housingData)
head(housing)
# fips county state time nSold medListPriceSqft medSoldPriceSqft
# 1 06001 Alameda County CA 2008-10-01 NA 307.9787 325.8118
# 2 06001 Alameda County CA 2008-11-01 NA 299.1667 NA
# 3 06001 Alameda County CA 2008-11-01 NA NA 318.1150
# 4 06001 Alameda County CA 2008-12-01 NA 289.8815 305.7878
# 5 06001 Alameda County CA 2009-01-01 NA 288.5000 291.5977
# 6 06001 Alameda County CA 2009-02-01 NA 287.0370 NAHousing Data Variables
| Variable | Description |
|---|---|
| fips | Federal Information Processing Standard - a 5 digit county code |
| county | US county name |
| state | US state name |
| time | date (the data is aggregated monthly) |
| nSold | number sold this month |
| medListPriceSqft | median list price per square foot (USD) |
| medSoldPriceSqft | median sold price per square foot (USD) |
Data Ingest
- When working with any data set, the first thing to do is create a ddo/ddf from the data
You Try It
Our housing data is already in memory, so we can use the simple approach of casting the data frame as a ddf:
library(datadr)
library(housingData)
housingDdf <- ddf(housing)# print result to see what it looks like
housingDdf
# Distributed data frame backed by 'kvMemory' connection
#
# attribute | value
# ----------------+----------------------------------------------------------------
# names | fips(fac), county(fac), state(fac), time(Dat), and 3 more
# nrow | 224369
# size (stored) | 10.58 MB
# size (object) | 10.58 MB
# # subsets | 1
#
# * Other attributes: getKeys()
# * Missing attributes: splitSizeDistn, splitRowDistn, summaryDivide by county and state
One useful way to look at the data is divided by county and state
byCounty <- divide(housingDdf,
by = c("county", "state"), update = TRUE)* update = TRUE tells datadr to compute and update the attributes of the ddf (seen in printout)
byCounty
# Distributed data frame backed by 'kvMemory' connection
#
# attribute | value
# ----------------+----------------------------------------------------------------
# names | fips(cha), time(Dat), nSold(num), and 2 more
# nrow | 224369
# size (stored) | 15.73 MB
# size (object) | 15.73 MB
# # subsets | 2883
#
# * Other attributes: getKeys(), splitSizeDistn(), splitRowDistn(), summary()
# * Conditioning variables: county, stateInvestigating the ddf
Recall that a ddf is a collection of key-value pairs
# look at first key-value pair
byCounty[[1]]
# $key
# [1] "county=Abbeville County|state=SC"
#
# $value
# fips time nSold medListPriceSqft medSoldPriceSqft
# 1 45001 2008-10-01 NA 73.06226 NA
# 2 45001 2008-11-01 NA 70.71429 NA
# 3 45001 2008-12-01 NA 70.71429 NA
# 4 45001 2009-01-01 NA 73.43750 NA
# 5 45001 2009-02-01 NA 78.69565 NA
# ...
Investigating the ddf
# look at first two key-value pairs
byCounty[1:2]
# [[1]]
# $key
# [1] "county=Abbeville County|state=SC"#
#
# $value
# fips time nSold medListPriceSqft medSoldPriceSqft
# 1 45001 2008-10-01 NA 73.06226 NA
# 2 45001 2008-11-01 NA 70.71429 NA
# 3 45001 2008-12-01 NA 70.71429 NA
# 4 45001 2009-01-01 NA 73.43750 NA
# 5 45001 2009-02-01 NA 78.69565 NA
# ...#
#
#
# [[2]]
# $key
# [1] "county=Acadia Parish|state=LA"#
#
# $value
# fips time nSold medListPriceSqft medSoldPriceSqft
# 1 22001 2008-10-01 NA 67.77494 NA
# 2 22001 2008-11-01 NA 67.16418 NA
# 3 22001 2008-12-01 NA 63.59129 NA
# 4 22001 2009-01-01 NA 65.78947 NA
# 5 22001 2009-02-01 NA 65.78947 NA
# ...Investigating the ddf
# lookup by key
byCounty[["county=Benton County|state=WA"]]
# $key
# [1] "county=Benton County|state=WA"
#
# $value
# fips time nSold medListPriceSqft medSoldPriceSqft
# 1 53005 2008-10-01 137 106.6351 106.2179
# 2 53005 2008-11-01 80 106.9650 NA
# 3 53005 2008-11-01 NA NA 105.2370
# 4 53005 2008-12-01 95 107.6642 105.6311
# 5 53005 2009-01-01 73 107.6868 105.8892
# ...You Try It
Try using the divide statement to divide on one or more variables
press down arrow key for solution when you're ready
Possible Solutions
byState <- divide(housing, by="state")
byMonth <- divide(housing, by="time")ddf Attributes
summary(byCounty)
# fips time nSold
# ------------------- ------------------ -------------------
# levels : 2883 missing : 0 missing : 164370
# missing : 0 min : 08-10-01 min : 11
# > freqTable head < max : 14-03-01 max : 35619
# 26077 : 140 mean : 274.6582
# 51069 : 140 std dev : 732.2429
# 08019 : 139 skewness : 10.338
# 13311 : 139 kurtosis : 222.8995
# ------------------- ------------------ -------------------
# ...
names(byCounty)
# [1] "fips" "time" "nSold" "medListPriceSqft"
# [5] "medSoldPriceSqft"
length(byCounty)
# [1] 2883
getKeys(byCounty)
# [[1]]
# [1] "county=Abbeville County|state=SC"
# ...Transformations
-
addTransform(data, fun) applies a function fun to each value of a key-value pair in the input ddo/ddf data
-
e.g. calculate a summary statistic for each subset
-
-
The transformation is not applied immediately, it is deferred until:
-
A function that kicks off a MapReduce job is called, e.g. recombine()
-
A subset of the data is requested (e.g. byCounty[[1]])
-
The drPersist() function explicitly forces transformation computation to persist
-
Transformation Example
Compute the slope of a simple linear regression of list price vs. time
# function to calculate a linear model and extract the slope parameter
lmCoef <- function(x) {
coef(lm(medListPriceSqft ~ time, data = x))[2]
}
# best practice tip: test transformation function on one subset
lmCoef(byCounty[[1]]$value)
# time
# -0.0002323686
# apply the transformation function to the ddf
byCountySlope <- addTransform(byCounty, lmCoef)Transformation example
# look at first key-value pair
byCountySlope[[1]]
# $key
# [1] "county=Abbeville County|state=SC"
#
# $value
# time
# -0.0002323686
# what does this object look like?
byCountySlope
# Transformed distributed data object backed by 'kvMemory' connection
#
# attribute | value
# ----------------+----------------------------------------------------------------
# size (stored) | 15.73 MB (before transformation)
# size (object) | 15.73 MB (before transformation)
# # subsets | 2883
#
# * Other attributes: getKeys()
# * Conditioning variables: county, state*note: this is a ddo (value isn't a data frame) but it is still pointing to byCounty's data
Exercise: create a transformation function
Create a new transformed data set using addTransform()
- Add/remove/modify variables
- Compute a summary (# rows, #NA, etc.)
- etc.
# get a subset
x <- byCounty[[1]]$value
# now experiment with x and stick resulting code into a transformation function
# some scaffolding for your solution:
transformFn <- function(x) {
## you fill in here
}
xformedData <- addTransform(byCounty, transformFn)
Hint: it is helpful to grab a subset and experiment with it and then stick that code into a function:
Possible Solutions
# example 1 - total sold over time for the county
totalSold <- function(x) {
sum(x$nSold, na.rm=TRUE)
}
byCountySold <- addTransform(byCounty, totalSold)
# example 2 - range of time for which data exists for county
timeRange <- function(x) {
range(x$time)
}
byCountyTime <- addTransform(byCounty, timeRange)
# example 3 - convert usd/sqft to euro/meter
us2eur <- function(x)
x * 2.8948
byCountyEur <- addTransform(byCounty, function(x) {
x$medListPriceMeter <- us2eur(x$medListPriceSqft)
x$medSoldPriceMeter <- us2eur(x$medSoldPriceSqft)
x
})
Note that the us2eur function was detected - this becomes useful in distributed computing when shipping code across many computers
Recombination
countySlopes <- recombine(byCountySlope, combine = combRbind)
Let's recombine the slopes of list price vs. time into a data frame.
This is an example of statistic recombination where the result is small and is pulled back into R for further investigation.
head(countySlopes)
# county state val
# Abbeville County SC -0.0002323686
# Acadia Parish LA 0.0019518441
# Accomack County VA -0.0092717711
# Ada County ID -0.0030197554
# Adair County IA -0.0308381951
# Adair County KY 0.0034399585Recombination Options
-
combine parameter controls the form of the result
-
combine=combRbind: rbind is used to combine all of the transformed key-value pairs into a local data frame in your R session - this is a frequently used option
-
combine=combCollect: transformed key-value pairs are collected into a local list in your R session
-
combine=combDdo: results are combined into a new ddo object
-
Others can be written for more sophisticated goals such as model coefficient averaging, etc.
-
Persisting a Transformation
us2eur <- function(x)
x * 2.8948
byCountyEur <- addTransform(byCounty, function(x) {
x$medListPriceMeter <- us2eur(x$medListPriceSqft)
x$medSoldPriceMeter <- us2eur(x$medSoldPriceSqft)
x
})
byCountyEur <- drPersist(byCountyEur)
Often we want to persist a transformation, creating a new ddo or ddf that contains the transformed data. Transforming and persisting a transformation is an analytic recombination.
Let's persist the US/sqft to EUR/sqmeter transformation using drPersist():
*note we can also use recombine(combine = combDdf) - drPersist() is for convenience
Joining Multiple Datasets
We have a few additional data sets we'd like to join with our home price data divided by county
head(geoCounty)
# fips county state lon lat rMapState rMapCounty
# 1 01001 Autauga County AL -86.64565 32.54009 alabama autauga
# 2 01003 Baldwin County AL -87.72627 30.73831 alabama baldwin
# 3 01005 Barbour County AL -85.39733 31.87403 alabama barbour
# 4 01007 Bibb County AL -87.12526 32.99902 alabama bibb
# 5 01009 Blount County AL -86.56271 33.99044 alabama blount
# 6 01011 Bullock County AL -85.71680 32.10634 alabama bullock
head(wikiCounty)
# fips county state pop2013 href
# 1 01001 Autauga County AL 55246 http://en.wikipedia.org/wiki/Autauga_County,_Alabama
# 2 01003 Baldwin County AL 195540 http://en.wikipedia.org/wiki/Baldwin_County,_Alabama
# 3 01005 Barbour County AL 27076 http://en.wikipedia.org/wiki/Barbour_County,_Alabama
# 4 01007 Bibb County AL 22512 http://en.wikipedia.org/wiki/Bibb_County,_Alabama
# 5 01009 Blount County AL 57872 http://en.wikipedia.org/wiki/Blount_County,_Alabama
# 6 01011 Bullock County AL 10639 http://en.wikipedia.org/wiki/Bullock_County,_AlabamaExercise: Divide by County and State
- To join these with our byCounty data set, we first must divide by county and state
- Give it a try:
byCountyGeo <- # ...
byCountyWiki <- # ...Exercise: Divide by County and State
byCountyGeo <- divide(geoCounty,
by=c("county", "state"))
byCountyWiki <- divide(wikiCounty,
by=c("county", "state"))byCountyGeo
# Distributed data frame backed by 'kvMemory' connection
#
# attribute | value
# ----------------+----------------------------------------------------------------
# names | fips(cha), lon(num), lat(num), rMapState(cha), rMapCounty(cha)
# nrow | 3075
# size (stored) | 8.14 MB
# size (object) | 8.14 MB
# # subsets | 3075
# ...byCountyGeo[[1]]
# $key
# [1] "county=Abbeville County|state=SC"
#
# $value
# fips lon lat rMapState rMapCounty
# 1 45001 -82.45851 34.23021 south carolina abbevilleJoining the Data
drJoin() takes any number of named parameters specifying ddo/ddf objects and returns a ddo of the objects joined by key
joinedData <- drJoin(
housing = byCountyEur,
slope = byCountySlope,
geo = byCountyGeo,
wiki = byCountyWiki)joinedData
# Distributed data object backed by 'kvMemory' connection
#
# attribute | value
# ----------------+----------------------------------------------------------------
# size (stored) | 29.23 MB
# size (object) | 29.23 MB
# # subsets | 3144
Joining the Data
What does this object look like?
str(joinedData[[1]])
# List of 2
# $ key : chr "county=Abbeville County|state=SC"
# $ value:List of 4
# ..$ housing:'data.frame': 66 obs. of 7 variables:
# .. ..$ fips : chr [1:66] "45001" "45001" "45001" "45001" ...
# .. ..$ time : Date[1:66], format: "2008-10-01" "2008-11-01" ...
# .. ..$ nSold : num [1:66] NA NA NA NA NA NA NA NA NA NA ...
# .. ..$ medListPriceSqft : num [1:66] 73.1 70.7 70.7 73.4 78.7 ...
# .. ..$ medSoldPriceSqft : num [1:66] NA NA NA NA NA NA NA NA NA NA ...
# .. ..$ medListPriceMetre: num [1:66] 1114 1078 1078 1120 1200 ...
# .. ..$ medSoldPriceMetre: num [1:66] NA NA NA NA NA NA NA NA NA NA ...
# ..$ slope : Named num -0.000232
# ..$ geo :'data.frame': 1 obs. of 5 variables:
# .. ..$ fips : chr "45001"
# .. ..$ lon : num -82.5
# .. ..$ lat : num 34.2
# .. ..$ rMapState : chr "south carolina"
# .. ..$ rMapCounty: chr "abbeville"
# ..$ wiki :'data.frame': 1 obs. of 3 variables:
# .. ..$ fips : chr "45001"
# .. ..$ pop2013: int 25007
# .. ..$ href : chr "http://en.wikipedia.org/wiki/Abbeville_County,_South Carolina"
# ...for a given county/state, the value is a list with elements according to data provided in drJoin()
Joined Data
Note that joinedData has more subsets than byCountyEur
length(byCountyEur)
# [1] 2883
length(joinedData)
# [1] 3144- Some of the data sets we joined have more counties than we have data for in byCountyEur
- drJoin() makes a subset for every unique key across all input data sets
- If data is missing in a subset, it doesn't have an entry
# Adams County Iowa only has geo and wiki data
joinedData[["county=Adams County|state=IA"]]$value
# $geo
# fips lon lat rMapState rMapCounty
# 1 19003 -94.70909 41.03166 iowa adams
#
# $wiki
# fips pop2013 href
# 1 19003 3894 http://en.wikipedia.org/wiki/Adams_County,_IowaBonus Problem
Can you write a transformation and recombination to get all counties for which joinedData does not have housing information?
Solution
noHousing <- joinedData %>%
addTransform(function(x) is.null(x$housing)) %>%
recombine(combRbind)
head(subset(noHousing, val == TRUE))
# county state val
# 2884 Adams County IA TRUE
# 2885 Adams County ND TRUE
# 2886 Antelope County NE TRUE
# 2887 Arthur County NE TRUE
# 2888 Ashley County AR TRUE
# 2889 Aurora County SD TRUEtx <- addTransform(joinedData, function(x) is.null(x$housing))
noHousing <- recombine(tx, combRbind)(without pipes)
Filtering
Suppose we only want to keep subsets of joinedData that have housing price data
- drFilter(data, fun) applies a function fun to each value of a key-value pair in the input ddo/ddf data
- If fun returns TRUE we keep the key-value pair
joinedData <- drFilter(joinedData,
function(x) {
!is.null(x$housing)
})# joinedData should now have as many subsets as byCountyAus
length(joinedData)
# [1] 2883Trelliscope
- We now have a ddo, joinedData, which contains pricing data as well as a few other pieces of information like geographic location and a link to wikipedia for 2,883 counties in the US
- Using this data, let's make a simple plot of list price vs. time for each county and put it in a Trelliscope display
Trelliscope Setup
- Trelliscope stores its displays in a "Visualization Database" (VDB), which is a collection of files on your disk that contain metadata about the displays
- To create and view displays, we must first establish a connection to the VDB
library(trelliscope)
# establish a connection to a VDB located in a directory "housing_vdb"
vdbConn("housing_vdb", name = "housingexample")- If this location has not already been initialized as a VDB, you will be prompted if you want to create one at this location (you can set autoYes = TRUE to skip the prompt)
- If it has been initialized, it will simply make the connection
Trelliscope Display Fundamentals
- A Trelliscope display is created with the function makeDisplay()
- At a minimum, a specification of a Trelliscope display must have the following:
- data: a ddo or ddf input data set
- name: the name of the display
- panelFn: a function that operates on the value of each key-value pair and produces a plot
- There are several other options, some of which we will see and some of which you can learn more about in the documentation
A Simple Panel Function
Plot list price vs. time using rbokeh
library(rbokeh)
timePanel <- function(x) {
xlim <- c("2008-07-01", "2014-06-01")
figure(ylab = "Price / Sq. Meter (EUR)",
xlim = as.Date(xlim)) %>%
ly_points(time, medListPriceMeter,
data = x$housing,
hover = list(time, medListPriceMeter))
}
# test it on a subset
timePanel(joinedData[[1]]$value)Make a Display
makeDisplay(joinedData,
name = "list_vs_time_rbokeh",
desc = "List price over time for each county.",
panelFn = timePanel
)
- This creates a display and adds its meta data to the directory we specified in vdbConn()
- We can view the display with this:
view()
# to go directly to this display
view("list_vs_time_rbokeh")Try Your Own Panel Function
Ideas: recreate the previous plot with your favorite plotting package, or try something new (histogram of prices, etc.)
Make a panel function,
test it,
and make a new display with it
Possible Solutions
library(lattice)
library(ggplot2)
latticeTimePanel <- function(x) {
xyplot(medListPriceMeter + medSoldPriceMeter ~ time,
data = x$housing, auto.key = TRUE,
ylab = "Price / Meter (EUR)")
}
makeDisplay(joinedData,
name = "list_vs_time_lattice",
desc = "List price over time for each county.",
panelFn = latticeTimePanel
)
ggTimePanel <- function(x) {
qplot(time, medListPriceMeter, data = x$housing,
ylab = "Price / Meter (EUR)") +
geom_point()
}
makeDisplay(joinedData,
name = "list_vs_time_ggplot2",
desc = "List price over time for each county.",
panelFn = ggTimePanel
)A Display with Cognostics
- The interactivity in these examples hasn't been too interesting
- We can add more interactivity through cognostics
- Like the panel function, we can supply a cognostics function that
- Takes a subset of the data as input and returns a named list of summary statistics or cognostics about the subset
- A helper function cog() can be used to supply additional attributes like descriptions to help the viewer understand what the cognostic is
- These can be used for enhanced interaction with the display, helping the user focus their attention on panels of interest
Cognostics Function
zillowCog <- function(x) {
# build a string for a link to zillow
st <- getSplitVar(x, "state")
ct <- getSplitVar(x, "county")
zillowString <- gsub(" ", "-", paste(ct, st))
# return a list of cognostics
list(
slope = cog(x$slope, desc = "list price slope"),
meanList = cogMean(x$housing$medListPriceMeter),
meanSold = cogMean(x$housing$medSoldPriceMeter),
lat = cog(x$geo$lat, desc = "county latitude"),
lon = cog(x$geo$lon, desc = "county longitude"),
wikiHref = cogHref(x$wiki$href, desc="wiki link"),
zillowHref = cogHref(
sprintf("http://www.zillow.com/homes/%s_rb/", zillowString),
desc="zillow link")
)
}# test it on a subset
zillowCog(joinedData[[1]]$value)
# $slope
# time
# -0.0002323686
#
# $meanList
# [1] 1109.394
#
# $meanSold
# [1] NaN
#
# $lat
# [1] 34.23021
#
# $lon
# [1] -82.45851
#
# $wikiHref
# [1] "<a href=\"http://en.wikipedia.org/wiki/Abbeville_County,_South Carolina\" target=\"_blank\">link</a>"
#
# $zillowHref
# [1] "<a href=\"http://www.zillow.com/homes/Abbeville-County-SC_rb/\" target=\"_blank\">link</a>"
Display with Cognostics
We simply add our cognostics function to makeDisplay()
makeDisplay(joinedData,
name = "list_vs_time_cog",
desc = "List price over time for each county.",
panelFn = timePanel,
cogFn = zillowCog
)Rencontres R 2016
By Ryan Hafen
Rencontres R 2016
- 4,822



