Omnibenchmark:
How to contribute a project?
Zürich,
14. & 16.12.22



Omnibenchmark is a platform for open and continuous community driven benchmarking
Method developer/
Benchmarker
Method user
Methods
Datasets



Metrics
Omnibenchmark
- continuous
- self-contained modules
- all "products" can be accessed
- provenance tracking
- anyone can contribute
A slightly different more realistic view
Benchmarker
contributor

GitLab

Workflow

Docker container

projects
orchestrator


Dataset

Dataset


Dataset

A slightly different more realistic view
contributer
user

omnibenchmark-python

omniValidator
benchmarker
projects

templates

omb-site

{
orchestrator

triplestore


omni-sparql
dashboards

how to contribute?
Each project is an independent module, that can interact with the remaining benchmark
flexible language, dependencies, code, ...
needs some general and benchmark specific rules
1. Start a new renku project

Hackathon namespace ?
Attribution!
Public
1. Start a new renku project
Template source repo
Available modules for any step!
Load
Branch name

1. Start a new renku project
Keyword that will be used by downstream projects to import this project
Important fields:
Should be the same for all components of your benchmark!
These information can always be changed later on

1. Start a new renku project
Jupyterlab or Rstudio session
Wait for the image to build
Make sure you are on the latest commit

project > new session
Always turn on
1. Start a new renku project

Configuration of your docker image, R packages, Py modules, etc.
Metadata to run the project
Scripts to insert your code
To modify:
1. Start a new renku project

Only file to run. Will use `config.yaml` and your script.
To run:
1. Start a new renku project
Step-by-step guide in the working document

2. Define your environment

TEMPLATE
Base image
Install R dependencies
Install python dependencies
2. Define your environment

install.R
requirements.txt

install optparse
Install omnibenchmark python
3. define your module
---
data:
name: "out_dataset"
title: "Output of an example OmniObject"
description: "describe module here"
keywords: ["example_dataset"]
script: "path/to/method/dataset/metric/script.py"
benchmark_name: "omni_celltype"
inputs:
keywords: ["import_this", "import_that"]
files: ["count_file", "dim_red_file"]
prefix:
count_file: "counts"
dim_red_file: ["features", "genes"]
outputs:
files:
corrected_counts:
end: ".mtx.gz"
meta:
end: ".json"
parameter:
names: ["param1", "param2"]
keywords: ["param_dataset"]
filter:
param1:
upper: 50
lower: 3
exclude: 12
file: "path/to/param/combinations/to/exclude.json"
src/config.yaml
TEMPLATE
General module infos
4. Provide your code
- Flexible language, code, etc.
- Minimal inputs and outputs are predefined depending on benchmark and module type! Check here!
- Automatic workflow generation, multiple parameter runs , updates etc.



Input types
output types
4. Provide your code:
R script arguments
### ----------------------------------------------------------------- ###
## ------------------------- Options ------------------------------- ##
### ----------------------------------------------------------------- ###
library(optparse)
option_list <- list(
make_option(c("--count_file"), type = "character", default = NULL,
help = "Path to count file (cells as columns, genes as rows)"),
make_option(c("--meta_file"), type = "character", default = NULL,
help = "Path to file with meta data infos")
);
opt_parser <- OptionParser(option_list = option_list);
opt <- parse_args(opt_parser);
if (is.null(opt$count_file) || is.null(opt$meta_file)) {
print_help(opt_parser)
stop("Please specify a path to the counts and meta data file.",
call. = FALSE)
}
count_file <- opt$count_file
meta_file <- opt$meta_file
method.R
module code
- Flexible language, code, etc.
- Minimal inputs and outputs are predefined depending on benchmark and module type! Check here!
- Automatic workflow generation, multiple parameter runs , updates etc.



Input types
output types
Omnibenchmark python module
- PyPI module to manage workflows and datasets
- Automation for new/unknown inputs
from omnibenchmark import get_omni_object_from_yaml, renku save
## Load config
omni_obj = get_omni_object_from_yaml('src/config.yaml')
## Check for new/updates of input datasets
omni_obj.update_object()
renku_save()
## Create output dataset
omni_obj.create_dataset()
## Generate and run workflow
omni_obj.run_renku()
renku_save()
## Store results in output dataset
omni_obj.update_result_dataset()
renku_save()5. Run omnibenchmark
from omnibenchmark.utils.build_omni_object import get_omni_object_from_yaml
from omnibenchmark.renku_commands.general import renku_save
## Load config
omni_obj = get_omni_object_from_yaml('src/config.yaml')
## Update object
omni_obj.update_object()
renku_save()
## Create output dataset
omni_obj.create_dataset()
## Run workflow
omni_obj.run_renku()
renku_save()
## Update Output dataset
omni_obj.update_result_dataset()
renku_save()run_workflow.py
TEMPLATE
5. Run omnibenchmark:
build object
from omnibenchmark.utils.build_omni_object import get_omni_object_from_yaml
from omnibenchmark.renku_commands.general import renku_save
## Load config
omni_obj = get_omni_object_from_yaml('src/config.yaml')
Warning: Skipped parameter because of missing information. Please specify at least names.
WARNING: No input files in the current project detected.
Run OmniObject.update_object() to update and import input datasets.
Otherwise check the specified prefixes: {'count_file': ['_counts'], 'meta_file': ['_meta']}
WARNING: No outputs/output file mapping in the current project detected.
Run OmniObject.update_object() to update outputs, inputs and parameter.
Currently this object can not be used to generate a workflow.
run_workflow.py
5. Run omnibenchmark:
update object
from omnibenchmark.utils.build_omni_object import get_omni_object_from_yaml
from omnibenchmark.renku_commands.general import renku_save
## Load config
omni_obj = get_omni_object_from_yaml('src/config.yaml')
## Update object
omni_obj.update_object()
renku_save()
run_workflow.py
5. Run omnibenchmark:
check object
## Check object
omni_obj.__dict__
{'name': 'filter_hvg',
'keyword': ['omni_clustering_filter'],
'title': 'Filtering based on the most variable genes.',
'description': '(Normalized) counts and reduced dimensions of filtered omni_clustering datasets, based on the top 10% most variable genes.',
'command': <omnibenchmark.core.output_classes.OmniCommand at 0x7f3478402820>,
'inputs': <omnibenchmark.core.input_classes.OmniInput at 0x7f3450edcb20>,
'outputs': <omnibenchmark.core.output_classes.OmniOutput at 0x7f3450ef5910>,
'parameter': None,
'script': 'src/filter_hvg.R',
'omni_plan': None,
'orchestrator': 'https://renkulab.io/knowledge-graph/projects/omnibenchmark/omni_clustering/orchestrator-clustering',
'benchmark_name': 'omni_clustering',
'wflow_name': 'filter_hvg',
'dataset_name': 'filter_hvg',
'renku': True,
'kg_url': 'https://renkulab.io/knowledge-graph'}run_workflow.py
5. Run omnibenchmark:
output dataset
## Create output dataset
omni_obj.create_dataset()
Renku dataset with name filter_hvg and the following keywords ['omni_clustering_filter'] was generated.
<Dataset c98a1b7958c6463bbea9fc2e314c6190 filter_hvg>run_workflow.py
5. Run omnibenchmark:
run workflow
## Run workflow
omni_obj.run_renku(all=False)
Loading required package: SummarizedExperiment
Loading required package: MatrixGenerics
Loading required package: matrixStats
...run_workflow.py
Stdout & Stderr --> console
5. Run omnibenchmark:
update output dataset
## Update Output dataset
omni_obj.update_result_dataset()
renku_save()run_workflow.py
6. Work with omnibenchmark:
Add metadata files to output dataset
from omnibenchmark.management.data_commands import update_dataset_files
### Update Output dataset
meta_files = [files["meta_file"] for files in omni_obj.inputs.input_files.values()]
update_dataset_files(urls=meta_files, dataset_name=omni_obj.name)
Added the following files to filter_hvg:
['data/koh/koh_meta.json', 'data/kumar/kumar_meta.json']
run_workflow.py
6. Work with omnibenchmark:
dirty repository
DirtyRepository: The repository is dirty. Please use the "git" command to clean it.
On branch master
Your branch is up to date with 'origin/master'.
Changes not staged for commit:
(use "git add <file>..." to update what will be committed)
(use "git restore <file>..." to discard changes in working directory)
modified: src/filter_hvg.R
no changes added to commit (use "git add" and/or "git commit -a")
Once you have added the untracked files, commit them with "git commit".Error
renku savefrom omnibenchmark.renku_commands.general import renku_save
renku_save()6. Work with omnibenchmark:
parameter
parameter:
names: ["order", "hvg", "k"]
keywords: ["omni_batch_parameter"]
default:
order: "auto"
hvg: "sig"
k: 20
filter:
k:
upper: 30
lower: 5
config.yaml
6. Work with omnibenchmark:
parameter
parameter:
names: ["param1", "param2"]
values:
param1: [10, 20]
param2: ["test", "another_test"]
combinations:
comb1:
param1: 10
param2: "test"
comb2:
param1: 20
param2: "another_test"
config.yaml
7. Add your module to an existing benchmark
Open an issue at the corresponding orchestrator

8. If you don't want to wait for renku to start a session/need more resources
Recipe to run omnibenchmark from the CLI



Next ...
- Sign up for a task here
- Pick a contribution - distribute within the group
- Discuss your setup
- Try it out!
Get started
- Hackathon document
- Read the Docs omnibenchmark
- Read the Docs renku
- Existing projects: omni_batch, omni_clustering
- Ask us!
Help
Omni-Batch-py:
Overview



What has been done?

- scib package
- AnnData objects of datasets
- scATAC integration (skip here)

- simulations
- methods/metrics

- Nextflow workflow
- methods/metrics

- batch mixing
- biological conservation
- simulations
Current state
Omni-Batch-py:
Overview



Omnibenchmark is a platform for open and continuous community driven benchmarking
Method developer/
Benchmarker
Method user
Methods
Datasets



Metrics
Omnibenchmark
- continuous
- self-contained modules
- all "products" can be accessed
- provenance tracking
- anyone can contribute
Omnibenchmark design
Data
standardized datasets


= 1 "module" (renku project )



Methods
method results

Metrics
metric results

Dashboard
interactive result exploration
Method user
Method developer/
Benchmarker
Omnibenchmark:
Workflows and activities


Omnibenchmark design
Data
standardized datasets


= 1 "module" (renku project )



Methods
method results

Metrics
metric results

Dashboard
interactive result exploration
Method user
Method developer/
Benchmarker
Omnibenchmark is a platform for open and continuous community driven benchmarking
Method developer/
Benchmarker
Method user
Methods
Datasets



Metrics
Omnibenchmark
- continuous
- self-contained modules
- all "products" can be accessed
- provenance tracking
- anyone can contribute
Omnibenchmark modules are independent and self-contained

GitLab project

Docker container

Workflow

Datasets
Collection of
method* history,
description how to run it, comp. environment, datasets
=
Workflows/Plans
- Workflow --> recipe how to generate outputs --> "plan"
- CWL --> open standard to describe workflows to make them portable and scalable across plattforms
- Workflow/plan != output generation



Input files
output files
How to generate workflows ?
make --> Makefile / snakemake --> snakefile
- define the tasks to be executed
- define the order of tasks
- define dependencies between tasks
--> user writes the "recipe" to a workflow file
renku run --name word_count -- wc < src/config.yaml > wc_config.txtWorkflow generation in renku
renku tracks a command as workflow:
- renku writes a recipe of what has been done
- translates the plan into triples
- tracks the plan and how it is used in form of triples



Input files
output files
Renku client is based on a Triplet store (Knowledge graph)

Result
Code
Data
generated
used_by
used_by
Data
Code
Result
used_by
generated
User interaction with renku client
Automatic triplet generation
Triplet store "Knowledge graph"
User interaction with renku client
KG-endpoint queries
Activities/Executions
- workflow executions/output generation
- One plan can be executed infinite times with the same and/or different inputs/outputs/parameter

How to execute activities?
make/snakemake --> make/snakemake
- generates all outputs that are not existing/uptodate
--> one command to automatically update and generate workflows without unnecessary reruns
renku run --name word_count -- wc < src/config.yaml > wc_config.txt
renku update --all
renku rerun wc_config.txt
renku workflow execute --set input-1="README.md" \
--set output-2="wc_readme.txt" \
word_countOmnibenchmark python
from omnibenchmark import get_omni_object_from_yaml, renku_save
## Load config
omni_obj = get_omni_object_from_yaml('src/config.yaml')
## Update object
omni_obj.update_object()
renku_save()
## Create output dataset
omni_obj.create_dataset()
## Run workflow
omni_obj.run_renku()
renku_save()
## Update Output dataset
omni_obj.update_result_dataset()
renku_save()renku workflows
Tricky with renku workflows
- changing/editing the recipe
- accessing activities
- output tracking
- readibility/workflow tracking from within a project
- automatization/ renku python api
Advantages of renku workflows
- workflow tracking/provenance
- accessing workflow executions from outside the project
- workflow portability (CWL)
Omnibenchmark:
Update and News


Science talk
Almut Lütge
30.11.2022
Agenda
Omnibenchmark website
- presentation of our new site
- set up
- feedback
Benchmark/Hackathon results
- bettr batch correction
- overview
Omnibenchmark site
- mkdocs
- renku project (dependent on utils project)
- mirrored to uzh gitlab
- hugo (go)
- uzh gitlab
- gitlab-ci.yml
Site generation

get_data.py

Gitlab-API
Triple store queries




datasets.qmd, projects.md,..



omnibenchmark.org



datasets.html,
project.html, ..



gitlab-ci.yml
variables:
HUGO_ENV: production
stages:
- build
- update
- render
image_build:
stage: build
image: docker:stable
services:
- docker:dind
script:
- echo $CI_REGISTRY_PASSWORD | docker login -u $CI_REGISTRY_USER $CI_REGISTRY \
--password-stdin
- docker build -t $CI_REGISTRY_IMAGE .
- docker push $CI_REGISTRY_IMAGE
gitlab-ci.yml
update_data:
stage: update
image:
name: $CI_REGISTRY_IMAGE:latest
before_script:
- git config --global user.name ${GITLAB_USER_NAME}
- git config --global user.email ${GITLAB_USER_EMAIL}
- url_host=$(git remote get-url origin | sed -e "s/https:\/\/gitlab-ci-token:.*@//g")
- git remote set-url --push origin "https://oauth2:${CI_PUSH_TOKEN}@${url_host}"
script:
- export GIT_PYTHON_REFRESH=quiet
- python3 src/data_gen/get_projects.py
- quarto render
- git status && git add . && git commit -m "Build site" && git status
- git push --follow-tags origin HEAD:$CI_COMMIT_REF_NAME -o ci.skip
gitlab-ci.yml
test_render:
stage: render
image: registry.gitlab.com/pages/hugo/hugo_extended:latest
before_script:
- apk add --update --no-cache git go
- hugo mod init gitlab.com/pages/hugo
script:
- hugo
rules:
- if: $CI_COMMIT_REF_NAME != $CI_DEFAULT_BRANCH
pages:
stage: render
image: registry.gitlab.com/pages/hugo/hugo_extended:latest
before_script:
- apk add --update --no-cache git go
- hugo mod init gitlab.com/pages/hugo
script:
- hugo
artifacts:
paths:
- public
rules:
- if: $CI_COMMIT_REF_NAME == $CI_DEFAULT_BRANCH
Omni-Batch-py

Omni-Batch-py

Omni-Batch-py Summary
- More methods/datasets/metrics to come ..
- Different ways to check results
- A working omnibenchmark
Back to metrics:
Summarized metrics - Food for thought


Current work
Almut Lütge
01.03.2023
scib-metrics
from scib_metrics.benchmark import Benchmarker
bm = Benchmarker(
adata,
batch_key="batch",
label_key="cell_type",
embedding_obsm_keys=["Unintegrated", "Scanorama", "LIGER", "Harmony"],
n_jobs=4,
)
bm.benchmark()Summarized results
bm.plot_results_table()
Summarized results
bm.plot_results_table(min_max_scale=False)


No scaling:
Min-max scaling:
No scaling:
Min-max scaling:




Why using Scaling?

- Metrics have different ranges/sensitivities
- How to summarize/represent metrics fairly?
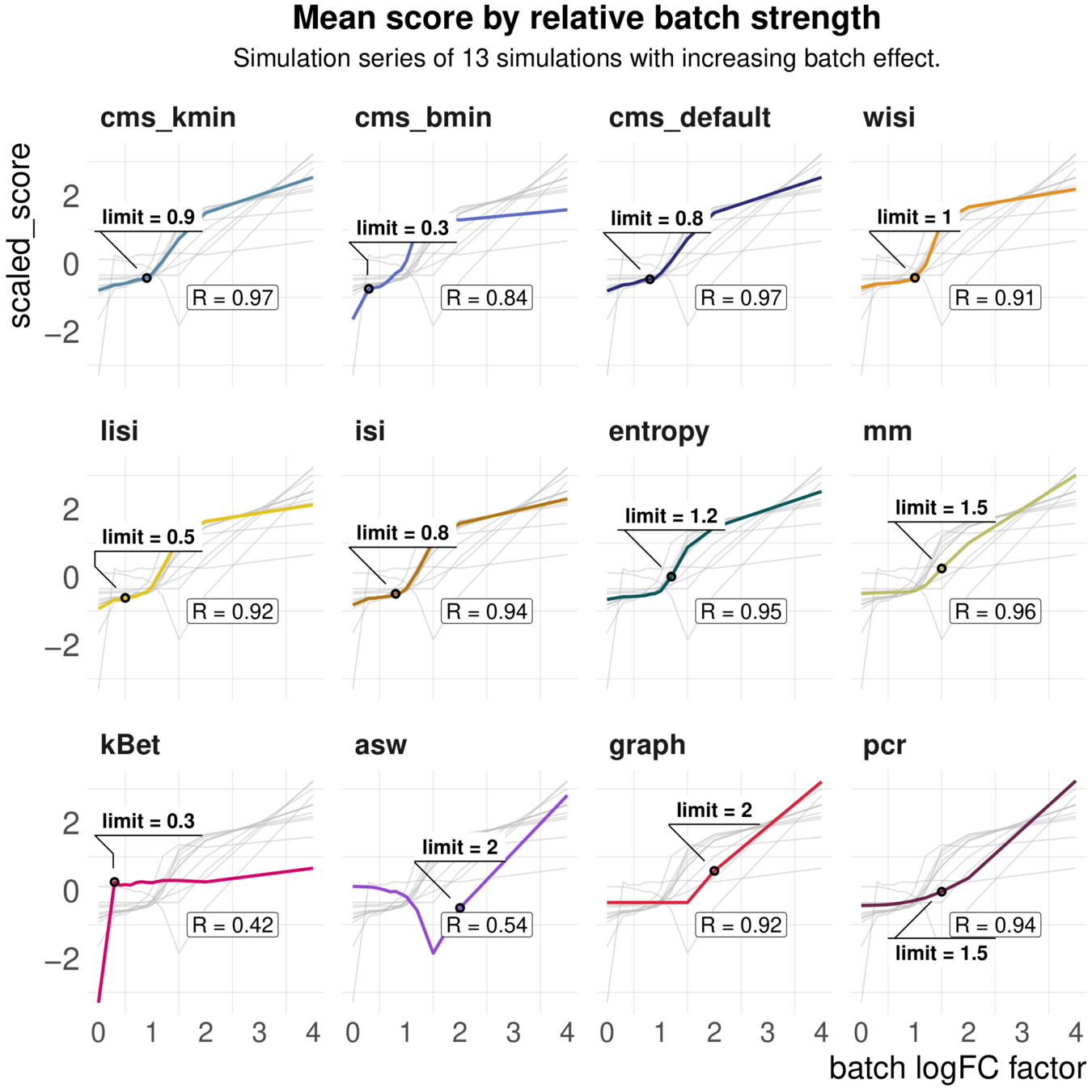
Metrics have different ranges to detect and distinguish batch effects
Larger range in results:
--> higher sensitivity
--> lower overall range
Another example
bm.plot_results_table()
Another example unscaled
bm.plot_results_table(min_max_scale=False)


No scaling:
Min-max scaling:
Antother example data
bm.get_results_table()
bettr?

Food for thought
Summaries
- Categorize data ? bettr to select limits?
- Use simulations to scale by metric range?
- Use metrics min/max?
To consider
- Should we include all metrics or only if they cover a new aspect?
- How to summarize within metric "groups"?

omni hackathon
By Almut Luetge
omni hackathon
- 138



